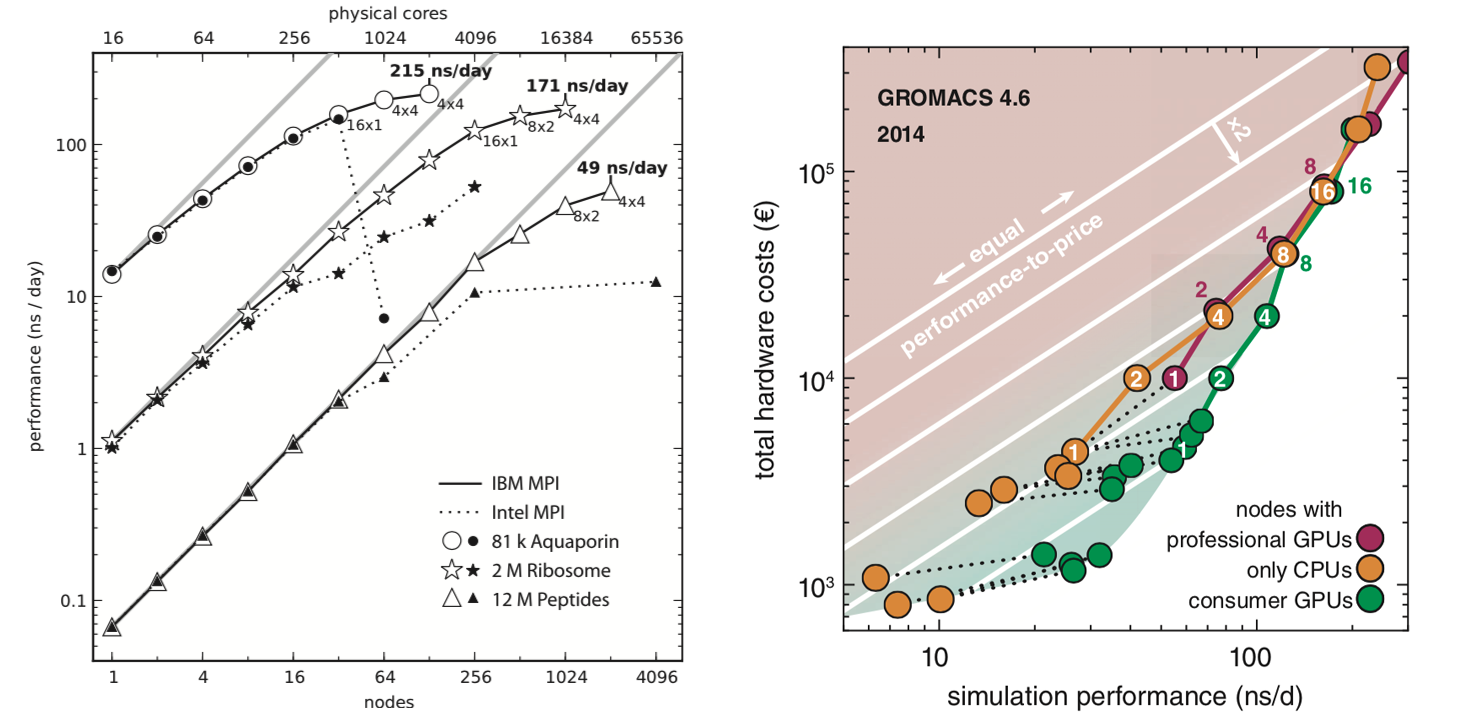
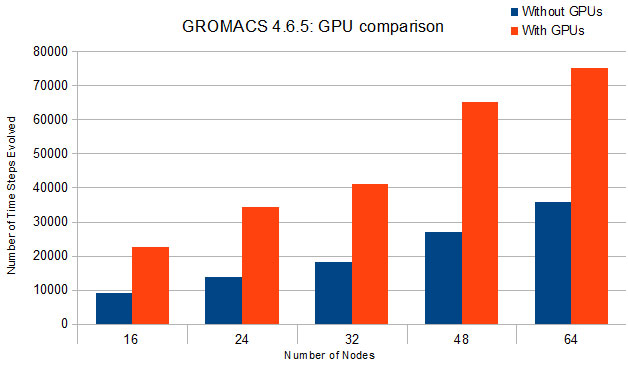
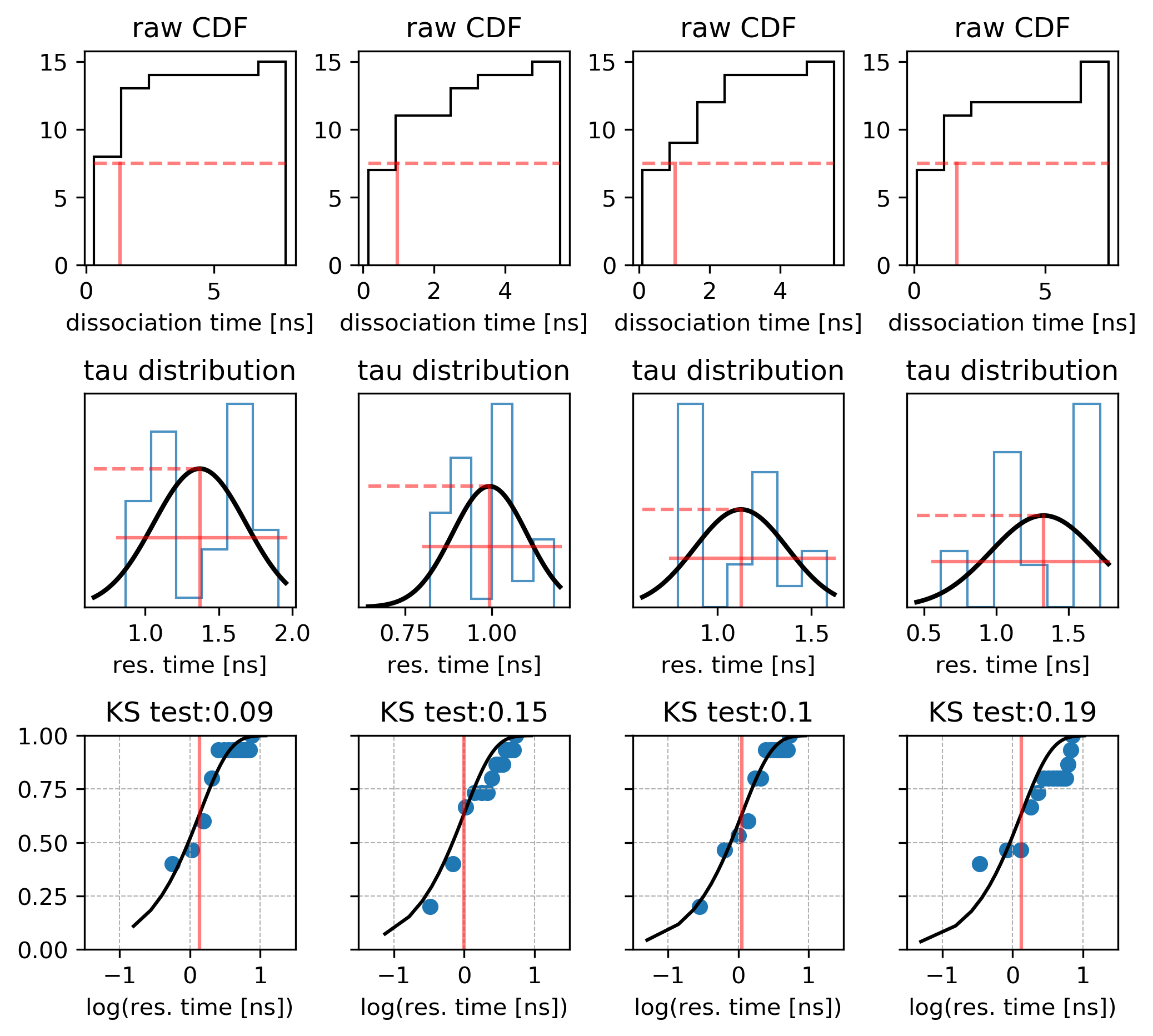
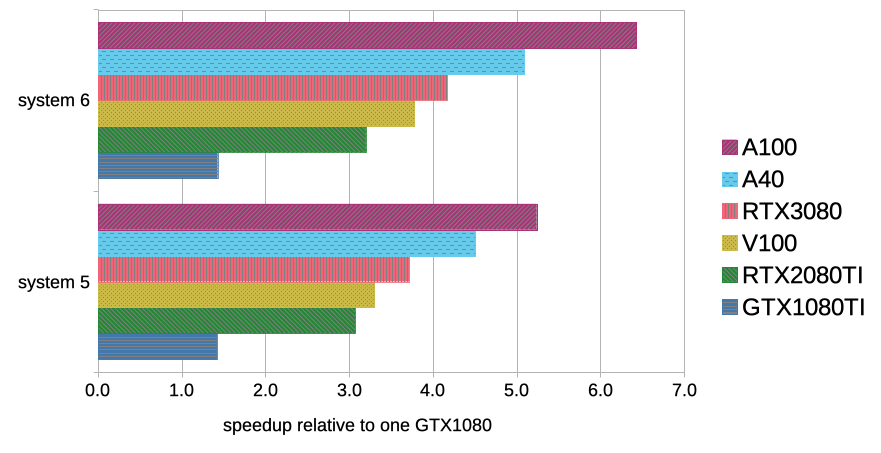
Using GPUs with Molecular Dynamics codes: optimizing usage from a user perspective Dr. Ole Juul Andersen CCK-11 February 1, ppt download

GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers - ScienceDirect