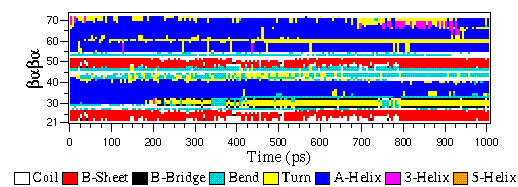
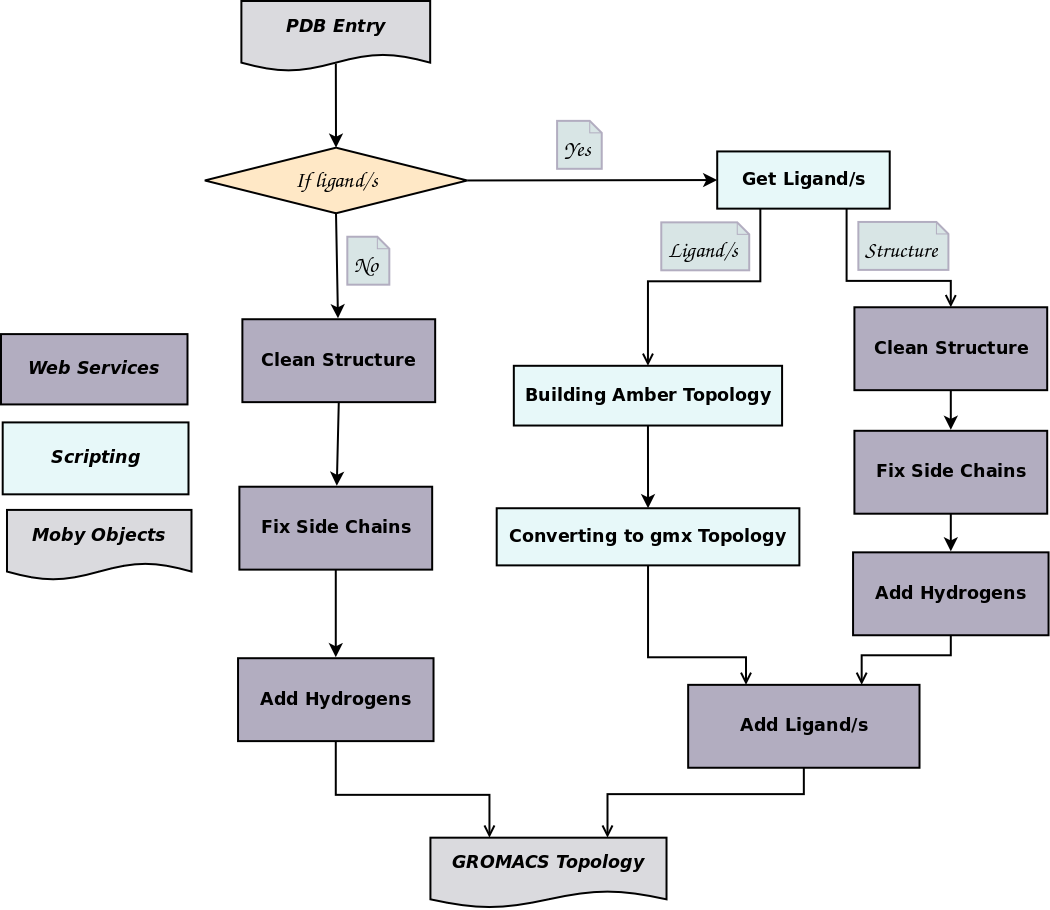
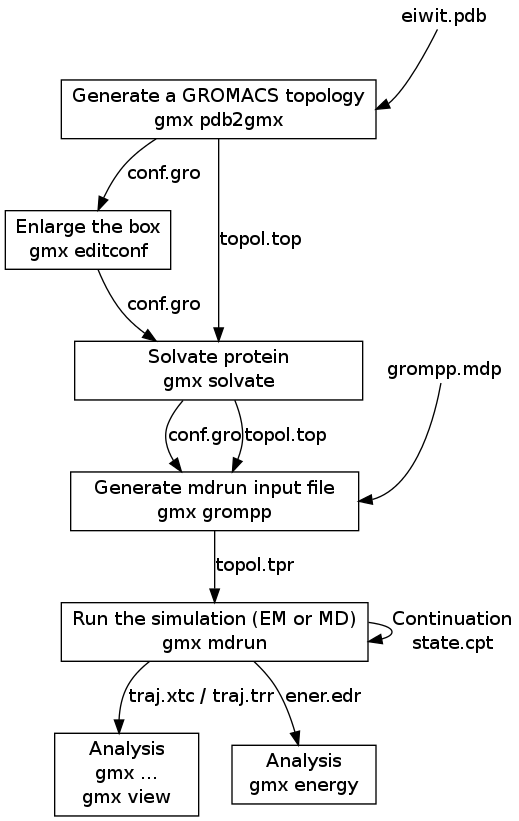
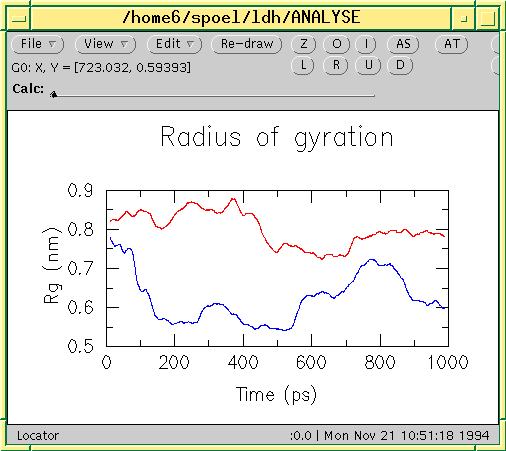
Data including GROMACS input files for atomistic molecular dynamics simulations of mixed, asymmetric bilayers including molecular topologies, equilibrated structures, and force field for lipids compatible with OPLS-AA parameters - ScienceDirect
Intuitive, reproducible high-throughput molecular dynamics in Galaxy: a tutorial | Journal of Cheminformatics | Full Text

How to build step wise the topology file for GROMACS (compatible with OPLS force field) for a newly designed ionic liquid?

RTP File Extension | Gromacs Residue Topology Parameter File | Associated Programs | Free Online Tools - FileProInfo